Monday, January 27
You can also download a PDF copy of this lecture.
The purpose of this exercise is to familiarize you with the
specification of linear models and contrasts within R. You will need to
have the following packages installed: abd,
cowplot, ggplot2, and
trtools. You should already have
ggplot2 and trtools installed. The
other packages can be installed using
install.packages("abd") and
install.packages("cowplot"), or
install.packages(c("abd","cowplot")).
This exercise features data from a study published in Nature
in 2006.1
The data can be found in the data frame MoleRats, included
in the package abd. You can inspect the data frame by
typing MoleRats at the prompt after you load the package
with library(abd), and you can look at a summary of the
data with summary(MoleRats) or the structure of the
variables with str(MoleRats). The study concerned two
“castes” of Damaraland mole-rats (Cryptomys damarensis):
frequent workers (denoted here as “worker”) and infrequent workers
(denoted here as “lazy”). The researchers observed the body mass (g) and
daily energy expenditure (kJ/day) of samples mole-rats from both castes.
Our objective is to model the difference between the two castes in terms
of daily energy expenditure. However, mole-rats in the two castes differ
by body mass, which may also be related to energy expenditure.
Here are plots of the raw data, showing the distribution of log energy expenditure for lazy and worker caste rats, and then again but also taking into account the (log) mass of the rats.
library(abd) # for the data
library(ggplot2) # for plotting
library(cowplot) # for use of the plot_grid function
# boxplot
p1 <- ggplot(MoleRats, aes(x = caste, y = ln.energy)) +
geom_boxplot() + geom_point() + theme_minimal() +
labs(x = "Caste", y = "Log Energy Expenditure (log kJ/Day)")
# scatterplot
p2 <- ggplot(MoleRats, aes(x = ln.mass, y = ln.energy, color = caste)) +
geom_point() + theme_minimal() +
theme(legend.position = "inside", legend.position.inside = c(0.9, 0.2)) +
labs(x = "Log Mass (log g)", y = NULL, color = "Caste")
# plot both plots side-by-side
plot_grid(p1, p2, align = "h", rel_widths = c(1,3))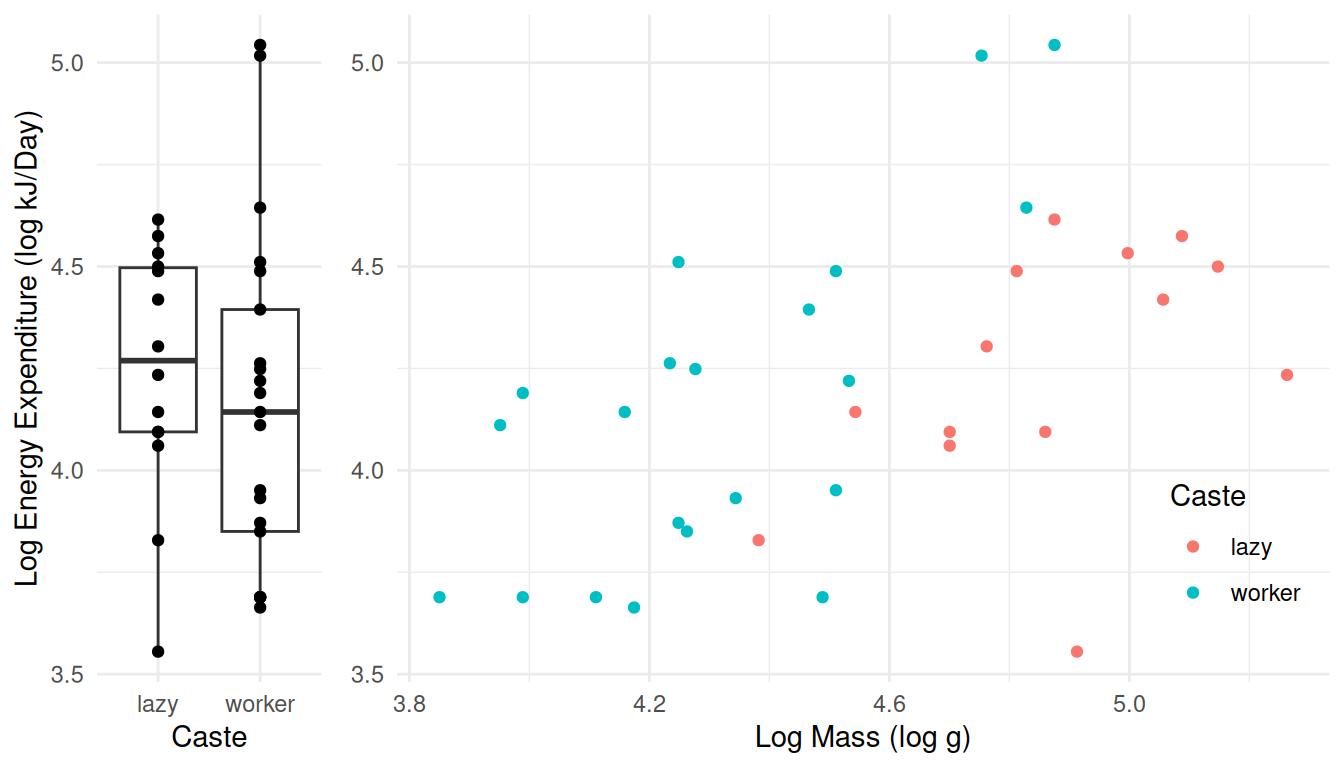 Note that both energy expenditure and mass are recorded on the (natural)
log scale. We will be modeling both of these variables on the log
scale.
Note that both energy expenditure and mass are recorded on the (natural)
log scale. We will be modeling both of these variables on the log
scale.
First consider a model where expected log energy expenditure is modeled as a function of only caste. Use the
lmfunction to estimate this model. We should see in the output of thesummaryfunction applied to the model object that R will, by default, parameterize this model as \[ E(Y_i) = \beta_0 + \beta_1 d_i, \] where \(Y_i\) is the log of energy expenditure of the \(i\)-th observation and \[ d_i = \begin{cases} 1, & \text{if the $i$-th observation is of a worker}, \\ 0, & \text{otherwise}, \end{cases} \] so that the model can also be written as \[ E(Y_i) = \begin{cases} \beta_0, & \text{if the $i$-th observation is from the lazy caste}, \\ \beta_0 + \beta_1, & \text{if the $i$-th observation is from the worker caste}. \end{cases} \]Solution: We can estimate the model as follows and confirm the parameterization using the output from
summary.m <- lm(ln.energy ~ caste, data = MoleRats) summary(m)$coefficientsEstimate Std. Error t value Pr(>|t|) (Intercept) 4.24601 0.1008 42.1150 2.885e-30 casteworker -0.08902 0.1302 -0.6839 4.988e-01The
casteworkershows that an indicator variable was created that assumes a value of one for any observation where the caste is worker, and zero otherwise.Consider the following three quantities and how they can be expressed as functions of the model parameters: the expected log energy expenditure of mole-rats from the lazy caste (\(\beta_0\)), the expected log energy expenditure of mole-rats from the worker caste (\(\beta_0 + \beta_1\)), and the difference in expected log energy expenditure between the two castes (\(\beta_1\) if we subtract lazy from worker). Note that using
summaryprovides inferences for \(\beta_0\) and \(\beta_1\), but not \(\beta_0 + \beta_1\). All three quantities can be written as a linear combination of the form \[ \ell = a_0\beta_0 + a_1\beta_1 + b. \] How can we use thelinconandcontrastfunctions to estimate these three quantities? Note that inferences for two of these quantities — i.e., the expected log expenditure of mole-rats of the lazy caste and the difference in the expected log expenditures between the two castes — should match those given by usingsummaryandconfintapplied to the model object.Solution: First we need to load the trtools package to use
linconandcontrast.library(trtools)We can use
linconto produce inferences for \(\beta_0\), \(\beta_0 + \beta_1\), and \(\beta_1\) as follows.lincon(m, a = c(1,0)) # b0estimate se lower upper tvalue df pvalue (1,0),0 4.246 0.1008 4.041 4.451 42.12 33 2.885e-30lincon(m, a = c(1,1)) # b0 + b1estimate se lower upper tvalue df pvalue (1,1),0 4.157 0.08232 3.99 4.324 50.5 33 7.89e-33lincon(m, a = c(0,1)) # b1estimate se lower upper tvalue df pvalue (0,1),0 -0.08902 0.1302 -0.3538 0.1758 -0.6839 33 0.4988Note, however, that single parameter inferences (i.e., \(\beta_0\) and \(\beta_1\)) are also produced by
summary. We can produce inferences about the expected response for the two castes usingcontrastas follows.contrast(m, a = list(caste = c("lazy","worker")), cnames = c("lazy","worker"))estimate se lower upper tvalue df pvalue lazy 4.246 0.10082 4.041 4.451 42.12 33 2.885e-30 worker 4.157 0.08232 3.990 4.324 50.50 33 7.890e-33And the following will produce inferences concerning the difference in the expected response between the worker and lazy castes.
contrast(m, a = list(caste = "worker"), b = list(caste = "lazy"))estimate se lower upper tvalue df pvalue -0.08902 0.1302 -0.3538 0.1758 -0.6839 33 0.4988Note that
linconanddcontrastproduce the same results. Also note that the estimated expected response for the worker caste is slightly less than that for the lazy caste, although the difference is not statistically significant at conventional significance levels.The model we considered above compares the two castes of mole-rats, but it does not account for differences in their sizes. The lazy mole-rats tend to be larger than the worker mole-rats. It may be useful to compare rats in the two castes while “controlling for size” meaning that comparisons can be made between mole-rats in different castes but of the same size. We can use
lmto estimate a linear model that includes both caste and the log of mass as explanatory variables. If we listcastefirst in the right-hand side of the model formula then the model will be parameterized as \[ E(Y_i) = \beta_0 + \beta_1 d_i + \beta_2 \log(m_i), \] where \(d_i\) is defined as before and \(\log(m_i)\) is the log of mass of the \(i\)-th observation (i.e., theln.massvariable). Note that this model can also be written as \[ E(Y_i) = \begin{cases} \beta_0 + \beta_2 \log(m_i), & \text{if the $i$-th observation is from the lazy caste}, \\ \beta_0 + \beta_1 + \beta_2 \log(m_i), & \text{if the $i$-th observation is from the worker caste}. \end{cases} \] Solution: The model can be estimated as follows.m <- lm(ln.energy ~ caste + ln.mass, data = MoleRats) summary(m)$coefficientsEstimate Std. Error t value Pr(>|t|) (Intercept) -0.09687 0.9423 -0.1028 9.188e-01 casteworker 0.39334 0.1461 2.6922 1.120e-02 ln.mass 0.89282 0.1930 4.6252 5.887e-05The parameterization can be confirmed from the output from
summary.Using the code given above, add lines to the plot to show the estimated expected log energy expenditure as a function of log mass and caste.
Solution: First we create a data set to hold estimated expected responses at a variety of values of the explanatory variables.
d <- expand.grid(caste = c("lazy","worker"), ln.mass = seq(3.8, 5.3, length = 100)) d$yhat <- predict(m, newdata = d)Then we “add to” the plot we created above.
p2 <- p2 + geom_line(aes(y = yhat), data = d) plot(p2)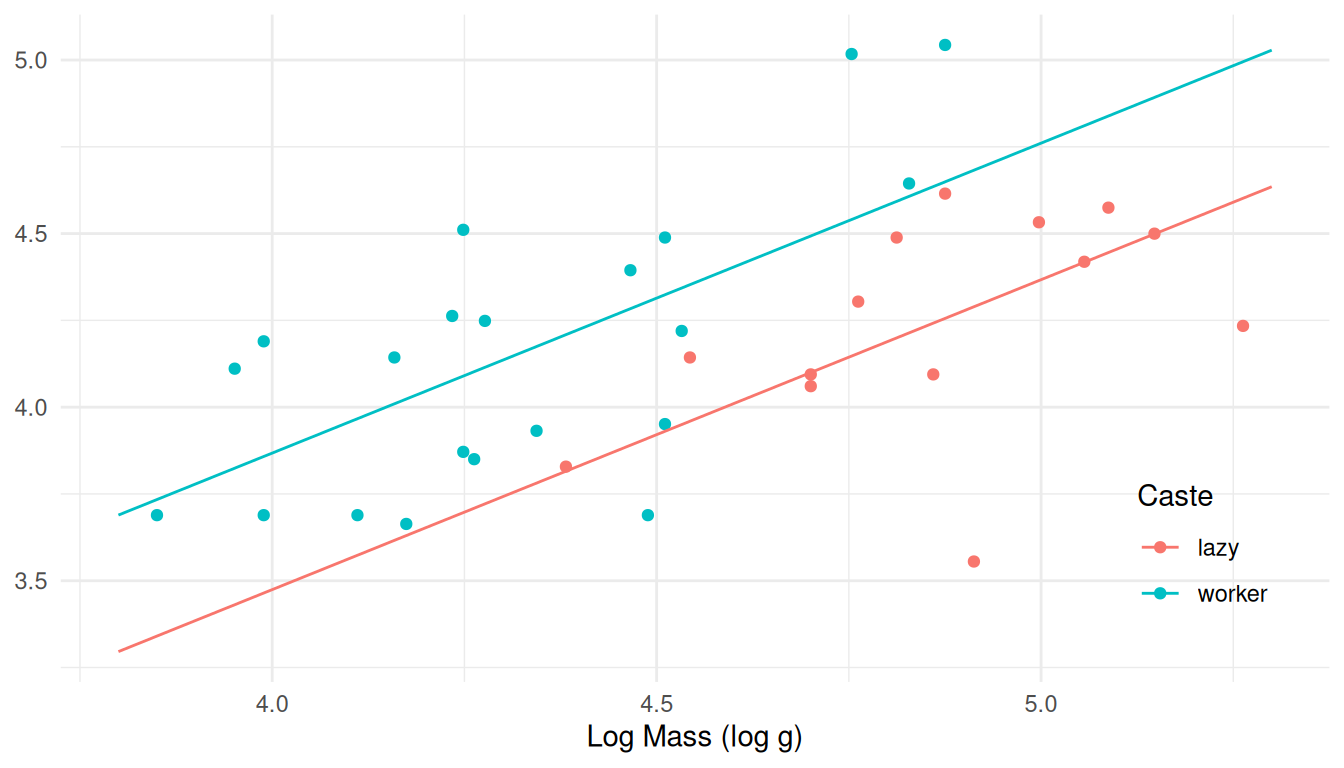 If we had not already created the plot object above then we could do as
follows.
If we had not already created the plot object above then we could do as
follows.p <- ggplot(MoleRats, aes(x = ln.mass, y = ln.energy, color = caste)) + geom_point() + theme_minimal() + theme(legend.position = "inside", legend.position.inside = c(0.9, 0.2)) + labs(x = "Log Mass (log g)", y = "Log Energy Expenditure (log kJ/Day)", color = "Caste") + geom_line(aes(y = yhat), data = d) plot(p)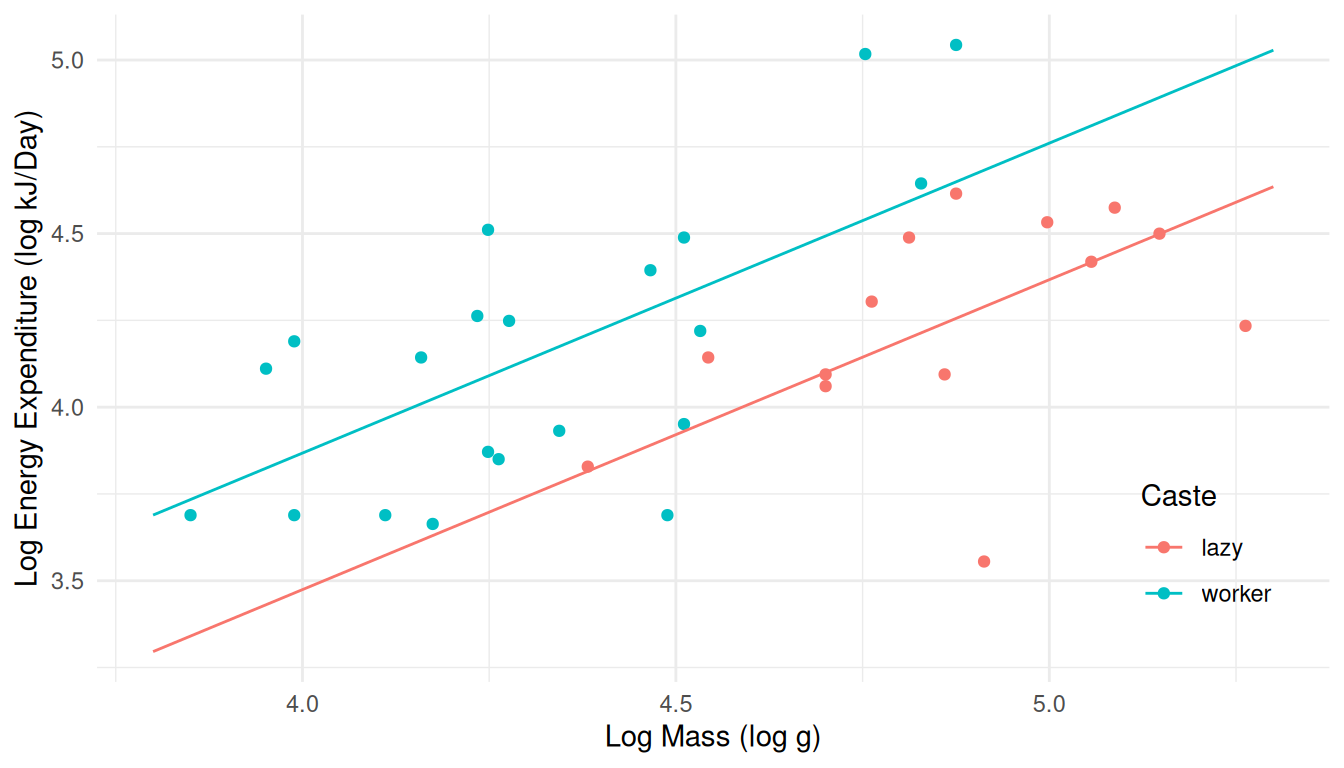
With the model now including the log of mass as an explanatory variable, comparisons between the castes must also consider mass. How can we use
contrastto estimate the expected log energy consumption for a mole-rat from each of the two castes with a log mass of 4.5? Also how can we estimate the difference in expected log energy expenditure between the two castes for a mole-rat with log mass 4.5? How can we do the same for log masses of 4.0 and 5.0? Our estimates should be consistent with the figure (i.e., they should look reasonable based on “eyeball” estimates from the figure). Also because the model specifies that the two lines are parallel, the estimated difference in expected log energy consumption between the castes will not depend on mass.Solution: Here are the estimates of the expected response for a lazy mole-rat at three different masses, and also a worker mole-rat at the same masses.worker
contrast(m, a = list(caste = "lazy", ln.mass = c(4,4.5,5)), cnames = c(4,4.5,5))estimate se lower upper tvalue df pvalue 4 3.474 0.18470 3.098 3.851 18.81 32 7.300e-19 4.5 3.921 0.10595 3.705 4.137 37.00 32 7.733e-28 5 4.367 0.08348 4.197 4.537 52.31 32 1.426e-32contrast(m, a = list(caste = "worker", ln.mass = c(4,4.5,5)), cnames = c(4,4.5,5))estimate se lower upper tvalue df pvalue 4 3.868 0.09000 3.684 4.051 42.98 32 7.067e-30 4.5 4.314 0.07309 4.165 4.463 59.02 32 3.120e-34 5 4.761 0.14566 4.464 5.057 32.68 32 3.725e-26And here we make inferences about the difference in the expected response at different values of log mass as follows.
contrast(m, a = list(caste = "worker", ln.mass = c(4,4.5,5)), b = list(caste = "lazy", ln.mass = c(4,4.5,5)), cnames = c(4,4.5,5))estimate se lower upper tvalue df pvalue 4 0.3933 0.1461 0.09573 0.691 2.692 32 0.0112 4.5 0.3933 0.1461 0.09573 0.691 2.692 32 0.0112 5 0.3933 0.1461 0.09573 0.691 2.692 32 0.0112The parameter \(\beta_2\) is the rate of change of the expected log energy consumption per unit increase in the log of mass. Inferences concerning this quantity can be obtained simply using
summaryandconfintapplied to the model object, but as an exercise how can we estimate this rate of change for each caste using thecontrastfunction? Naturally we should find that the estimated rate is the same for each caste.Solution: Here is how to use
contrastto estimate the rate of change in the expected response per unit increase in log mass for each caste.contrast(m, a = list(caste = c("lazy","worker"), ln.mass = 3), b = list(caste = c("lazy","worker"), ln.mass = 2), cnames = c("lazy","worker"))estimate se lower upper tvalue df pvalue lazy 0.8928 0.193 0.4996 1.286 4.625 32 5.887e-05 worker 0.8928 0.193 0.4996 1.286 4.625 32 5.887e-05A test to determine if there is a statistically significant difference in expected log energy expenditure between the two castes uses the null hypothesis \(H_0\!: \beta_1 = 0\) based on how the model was parameterized earlier. How can we test this null hypothesis using the full-null model approach (i.e., by specifying a null model based on the full model but with \(\beta_1 = 0\)) and using the
anovafunction? We should find that the resulting \(F\) test statistic is the square of the \(t\) test statistic fromsummary, and that the p-values fromsummaryandanovaare the same.Solution: Here is how to conduct the test using
anova.m.null <- lm(ln.energy ~ ln.mass, data = MoleRats) anova(m.null,m)Analysis of Variance Table Model 1: ln.energy ~ ln.mass Model 2: ln.energy ~ caste + ln.mass Res.Df RSS Df Sum of Sq F Pr(>F) 1 33 3.45 2 32 2.81 1 0.637 7.25 0.011 * --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Repeat 3-6 with both
casteandln.massas explanatory variables, but now with an “interaction” between the them by includingcaste:ln.energyin your model formula. Note that this model can be written case-wise as \[ E(Y_i) = \begin{cases} \beta_0 + \beta_2 \log(m_i), & \text{if the $i$-th observation is from the lazy caste}, \\ \beta_0 + \beta_1 + (\beta_2 + \beta_3) \log(m_i), & \text{if the $i$-th observation is from the worker caste}. \end{cases} \] Solution: The model would be specified asln.energy ~ caste + ln.mass + caste:ln.mass. Here are the parameter estimates and a plot of the estimated model.m <- lm(ln.energy ~ caste + ln.mass + caste:ln.mass, data = MoleRats) summary(m)$coefficientsEstimate Std. Error t value Pr(>|t|) (Intercept) 1.2939 1.6691 0.7752 0.44408 casteworker -1.5713 1.9518 -0.8050 0.42694 ln.mass 0.6069 0.3428 1.7706 0.08646 casteworker:ln.mass 0.4186 0.4147 1.0094 0.32061d <- expand.grid(caste = c("lazy","worker"), ln.mass = seq(3.8, 5.3, length = 100)) d$yhat <- predict(m, newdata = d) p <- ggplot(MoleRats, aes(x = ln.mass, y = ln.energy, color = caste)) + geom_point() + theme_classic() + theme(legend.position = "inside", legend.position.inside = c(0.9, 0.2)) + geom_line(aes(y = yhat), data = d) + labs(x = "Log Mass (log g)", y = "Log Energy Expenditure (log kJ/Day)", color = "Caste") plot(p)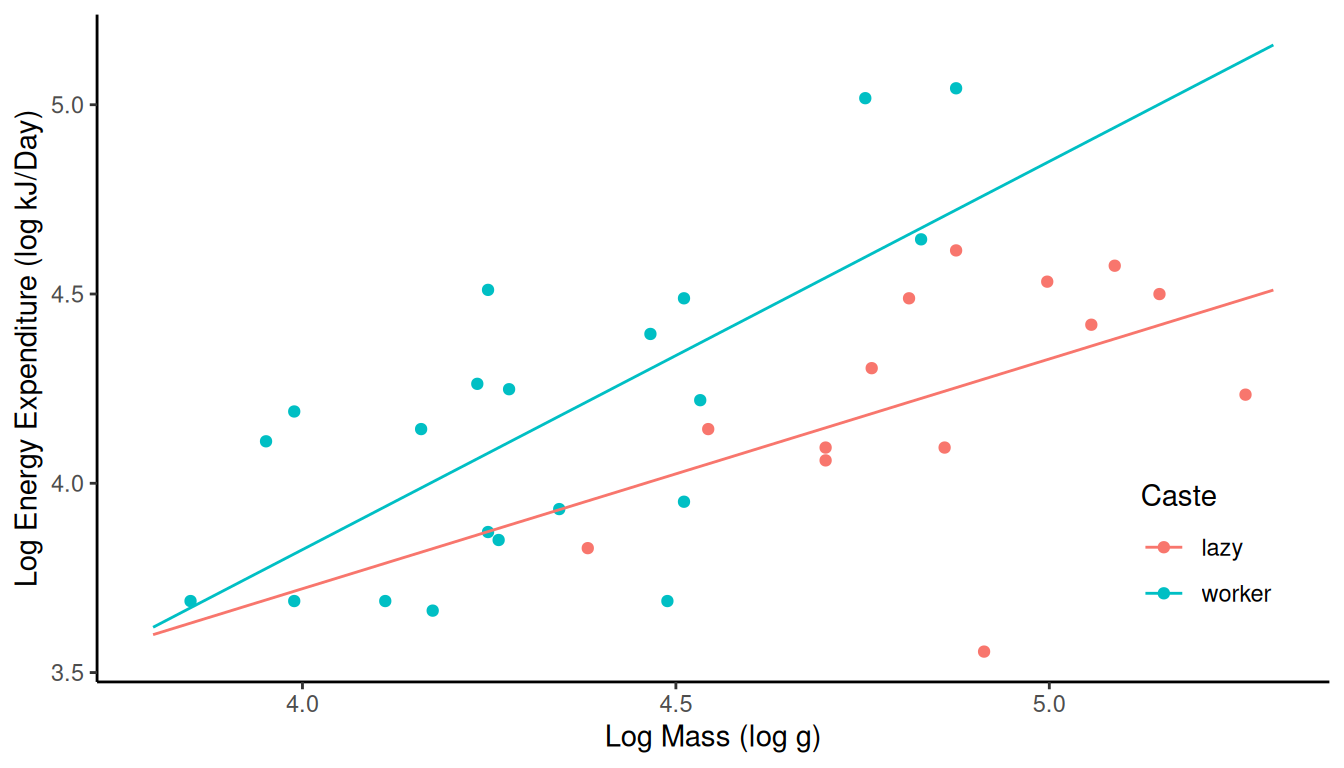 Notice how the “interaction” between log mass and caste now changes the
results form
Notice how the “interaction” between log mass and caste now changes the
results form contrastsince the lines are no longer restricted to be parallel.contrast(m, a = list(caste = "lazy", ln.mass = c(4,4.5,5)), cnames = c(4,4.5,5))estimate se lower upper tvalue df pvalue 4 3.722 0.30664 3.096 4.347 12.14 31 2.601e-13 4.5 4.025 0.14787 3.723 4.327 27.22 31 3.313e-23 5 4.328 0.09189 4.141 4.516 47.10 31 2.057e-30contrast(m, a = list(caste = "worker", ln.mass = c(4,4.5,5)), cnames = c(4,4.5,5))estimate se lower upper tvalue df pvalue 4 3.825 0.09954 3.622 4.028 38.42 31 1.026e-27 4.5 4.337 0.07665 4.181 4.494 56.59 31 7.432e-33 5 4.850 0.17059 4.502 5.198 28.43 31 9.024e-24contrast(m, a = list(caste = "worker", ln.mass = c(4,4.5,5)), b = list(caste = "lazy", ln.mass = c(4,4.5,5)), cnames = c(4,4.5,5))estimate se lower upper tvalue df pvalue 4 0.1032 0.3224 -0.55429 0.7608 0.3202 31 0.75095 4.5 0.3125 0.1666 -0.02715 0.6522 1.8765 31 0.07002 5 0.5219 0.1938 0.12667 0.9171 2.6932 31 0.01131contrast(m, a = list(caste = c("lazy","worker"), ln.mass = 3), b = list(caste = c("lazy","worker"), ln.mass = 2), cnames = c("lazy","worker"))estimate se lower upper tvalue df pvalue lazy 0.6069 0.3428 -0.09217 1.306 1.771 31 0.0864587 worker 1.0255 0.2335 0.54928 1.502 4.392 31 0.0001216
Note that for these both the response variable and one explanatory variable have been transformed using a log transformation. Suppose that the transformations had not been yet applied. To simulate this I will create two new variables which are the un-transformed variables.
MoleRats$energy <- exp(MoleRats$ln.energy)
MoleRats$mass <- exp(MoleRats$ln.mass)
summary(MoleRats) caste ln.mass ln.energy energy mass
lazy :14 Min. :3.85 Min. :3.56 Min. : 35.0 Min. : 47
worker:21 1st Qu.:4.25 1st Qu.:3.90 1st Qu.: 49.5 1st Qu.: 70
Median :4.51 Median :4.19 Median : 66.0 Median : 91
Mean :4.54 Mean :4.19 Mean : 71.0 Mean :100
3rd Qu.:4.84 3rd Qu.:4.49 3rd Qu.: 89.0 3rd Qu.:127
Max. :5.26 Max. :5.04 Max. :155.0 Max. :193 Note that \(\exp(x)\) is the inverse transformation of the natural logarithm. If this was the case then we can specify the transformations within the model formula itself.
m <- lm(log(energy) ~ caste + log(mass) + caste:log(mass), data = MoleRats)
summary(m)$coefficients Estimate Std. Error t value Pr(>|t|)
(Intercept) 1.2939 1.6691 0.7752 0.44408
casteworker -1.5713 1.9518 -0.8050 0.42694
log(mass) 0.6069 0.3428 1.7706 0.08646
casteworker:log(mass) 0.4186 0.4147 1.0094 0.32061Of course we obtain the same results. But when we make our plot we can put the explanatory variable on its original scale.
d <- expand.grid(caste = c("lazy","worker"), mass = seq(47, 193, length = 100))
d$yhat <- predict(m, newdata = d)
p <- ggplot(MoleRats, aes(x = mass, y = log(energy), color = caste)) +
geom_point() + theme_minimal() +
theme(legend.position = "inside", legend.position.inside = c(0.9, 0.2)) +
geom_line(aes(y = yhat), data = d) +
labs(x = "Mass (g)", y = "Log Energy Expenditure (log kJ/Day)", color = "Caste")
plot(p)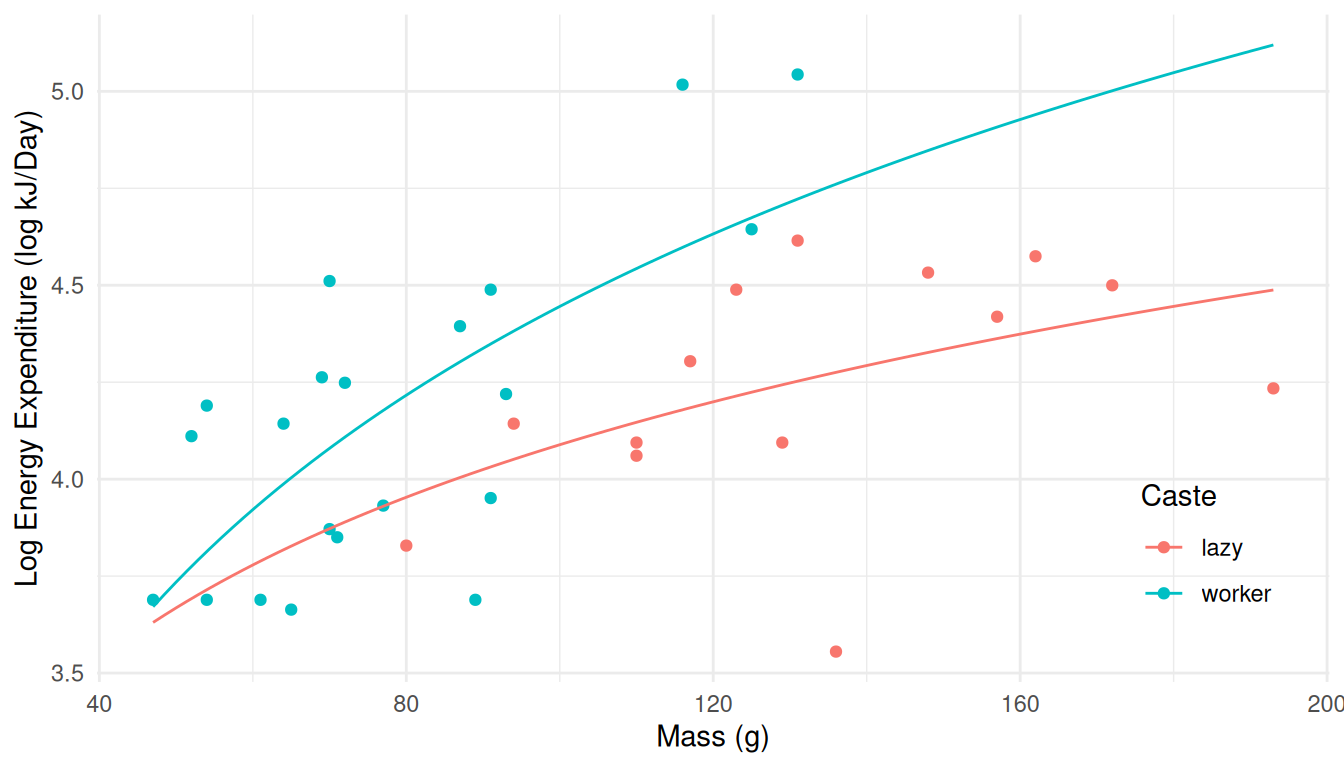 This shows that although this is a linear model, the expected response
(the logarithm of the energy expenditure) is not a linear
function of mass. And while we could use energy expenditure instead of
its logarithm on the ordinate for plotting the data, this does not make
sense for the data because we are modeling the expected value of the
logarithm of energy expenditure. You cannot “undo” a nonlinear
transformation of the response variable because it is not true that
\(E[f(x)] = f[E(x)]\) where \(f(x)\) is some nonlinear function (like
log).
This shows that although this is a linear model, the expected response
(the logarithm of the energy expenditure) is not a linear
function of mass. And while we could use energy expenditure instead of
its logarithm on the ordinate for plotting the data, this does not make
sense for the data because we are modeling the expected value of the
logarithm of energy expenditure. You cannot “undo” a nonlinear
transformation of the response variable because it is not true that
\(E[f(x)] = f[E(x)]\) where \(f(x)\) is some nonlinear function (like
log).
Scantlebury, M., Speakman, J. R., Oosthuizen, M. K., Roper, T. J., & Bennett, N. C. (2006). Energetics reveals physiological distinct castes in a eusocial mammal. Nature, 440, 795-797.↩︎